Author, developer and maintianer is: Obada Al Zoubi. LinkedIn and Personal Website
If you have requests, email me at: obada.y.alzoubi@gmail.com
Please note that the website is under development. Demos, tutorials will be added subsequently
The Toolbox:
{HFeSNN} is matlab toolbox for implementing Evolving Spiking Neural Networks (eSNNs) and Hierarchical Fusion eSNN (HFeSNN)
I developed this toolbox during my Ph.D. work in order to come up with a new class of ML approach for challenging problems. I made the algorithm and the toolbox open source so it can benefit the general public. Currently, the toolbox is targeting academic interest with potential industrial applications.Why Evolving Learning?
A majority of machine learning (ML) approaches functions in offline or batch modes, which limits their application to adaptive environments. Thus, developing algorithms that work in adaptive and dynamic environments is the subject of ongoing research. Such algorithms require to learn not only from new samples (online learning), but also from novel and unseen before knowledge.Here, the toolbox uses evolving learning (EL) to refer to learning from new knowledge and unseen-before classes without needing to re-train models as in traditional ML methods.
To achieve the goal of EL, we adopt a biologically-inspired paradigm to build a highly adaptive supervised learning algorithm based on two brain-like information processing: divide-and-conquer and hierarchical abstraction.Furthermore, our proposed algorithm, which we named it as Hierarchical Fusion Evolving Spiking Neural Network (HFSNN), uses a dynamical and biologically inspired spiking neural network (SNN) with the optimized neural model.
HFSNN does not impose any limitation on the data regarding the number of classes or the way of feeding the data to the model. The toolbox implements several features including learning in offline, online and evolving learning mods and establish for future applications for EL.
Depiction of the algorithm

Note the toolbox is highly customizable and targets development and building eSNN
Currently, HFeSNN supports classification only.
Installation
In Matlab, run
% clc;clear
% Add path for some needed codes
addpath('matlab')
addpath('data')
addpath('coreCode')Feature Encoding
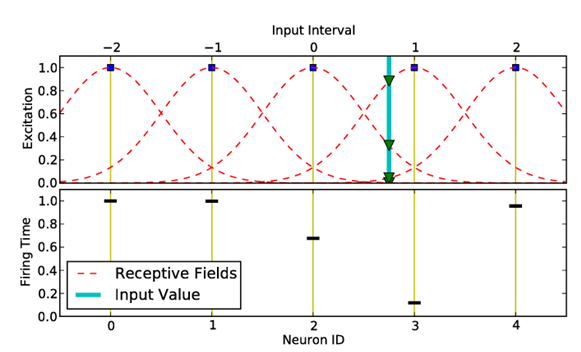
The rank coding procedure is the following: first, it starts by converting the input values into a sequence of spikes using the Gaussian receptive fields. The Gaussian receptive fields consist of m receptive fields that are used to represent the input value n into spikes. Assume that n takes values from the range [I_min^n ,I_max^n], then the Gaussian receptive field of neuron I is given by its center u_i:
HeSNN uses population encoding to convert features into set of spikes. The goal is to convert each feature to nbfields
neurons.
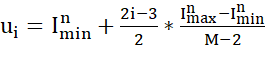
with

HeSNN uses matrix format to represent the data with last column is the class label. The features should be scaled between -1 and +1 in order to have correct encoding for the features.
-
Data: A matrix(nxm), with m column being corresponding labels. -
Param: A structure with several fields to control the training process.-
Beta:the width of the -
I_min:the lower level of receptive filed. -
I_max:the upper level of receptive filed. -
nbfields:the number of receptive fields per feature. -
max_response_threshold:firing threshold for neurons. -
s:merging threshold for neurons. -
m:exponential base decaying factors for neurons. -
c:discounting threshold for neurons. -
distance:the distance measure to merge neurons. -
distribution:the distribution for the receptive filed -
Beta:the width of the
-
Demo Example
In this demo example, we will use fisheriris dataset as an exmaple:
Al Zoubi, Obada, et al. “Hierarchical Fusion Evolving Spiking Neural Network for Adaptive Learning.” 2018 IEEE 17th International Conference on Cognitive Informatics & Cognitive Computing (ICCI* CC). IEEE, 2018.
dist = 'euclidean';% distance metric to merge neurons
pdf_f = 'Normal';% Experimetnal .. use Normal for default. Other receptive fields can be used.
s = 0.1;% threshold of merging neurons. High value will merge more neurons and make model more simple
c = 0.8; % satuartion controling variable
m = 0.9;% m paramter for Thorp's neural model ..in case you are using Thorp's model. if you are using NRO model presented in the paper (See refernces), you don't need m paramter.
I_min = -1; % lower range of the receptive field
I_max = +1; % upper range of the receptive field
nbfields = 32; % number of neurons to reprent each feature
Beta= 1.5; % the width of guassain field The previous parmaters should be provided as data strcture as following :
Param.m=m;
Param.c=c;
Param.s=s;
Param.pdf_option = pdf_f;
Param.pd_f = pdf_f;
Param.dist = dist;
max_response_time = 0.9;
% See Equation 1 in the paper for more infromation about I_min and I_max
Param.I_min=I_min;
Param.I_max=I_max;
% nbfields is M in the paper. The number of neurons to reperesnt each feature
Param.nbfields=nbfields;
% Beta paramters in equation 2 of the manuscript
Param.Beta=Beta;
Param.useVal = 0;% use validation in trainign ( not used now)
% Experimetnal
Param.eval =1000; % evaluate training ever specefic number ( not used now)
Param.useThreshold = true; % don't worry about this
Param.useClassWideRespose = true; % don't worry about this
Param.max_response_time = max_response_time; % don't worry about thisNow, let's load the fisheriris dataset as following:
%% fisheriris dataset
load fisheriris
irislas=ones(150,1);
irislas(51:100)=irislas(51:100)*2;
irislas(101:150)=irislas(101:150)*3;
Data=normalize(meas(:,1:2));
Data=[Data irislas];For training, simply:
repos =train_eSNN4(Dat_train, Param);The training function returns repository, repo, which is similar to the trained model in classifcal machine learning algorithms. The differnce is that repo structure can be flexible for learning from new and unseen classes. Also, it can evlove to new information, if needed.
For testing, simply:
%% fisheriris dataset
[Accuracy,predicted_labels]=test_eSNN3(Dat_test ,Param );The testing function return the classifcation accuracy and predicted labels, as in the tradiational machine learning algorithms.
Bug report
Please open an issue if you find a bug.
I will always redirect you to GitHub issues if you email me, so that others can benefit from our discussion.